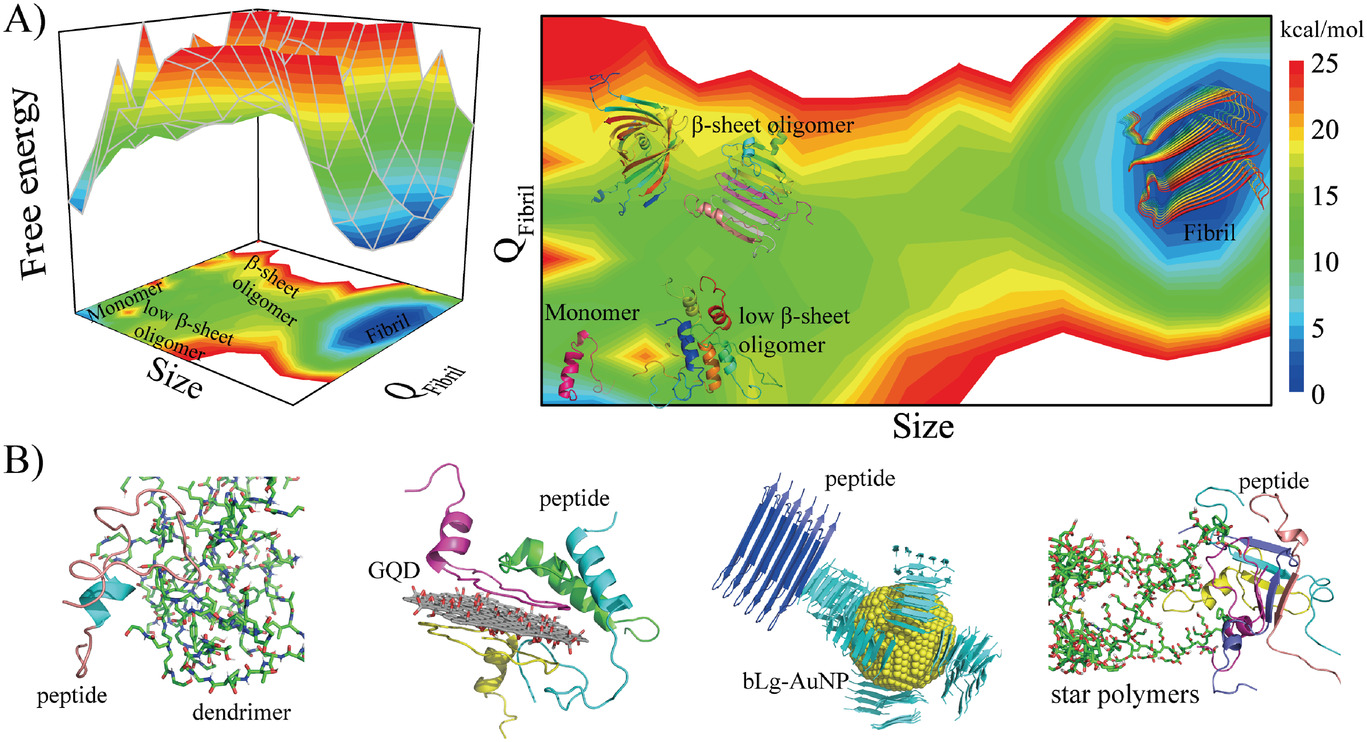
Amyloid aggregation is associated with an increasing list of degenerative diseases such as Alzheimer’s disease, Parkinson’s disease, and type-2 diabetes. The hallmark common to all amyloid diseases is the deposition of beta-sheet rich fibrils formed of by misfolding and aggregation of otherwise soluble proteins and peptides. Mounting experiments suggest that soluble oligomers instead of mature fibrils are more toxic. As the intrinsically transient and heterogenous intermediates of amyloid aggregation, these oligomeric species are highly elusive. In addition, not all oligomers are toxic. The objective of our research is to uncover the structure and dynamics of amyloid aggregation and focus on the early aggregation process in order to pinpoint the toxic subspecies. In our lab, we apply multiscale molecular dynamics simulations to model the aggregation of various amyloidogenic proteins and peptides, characterize the free energy landscape of the assembly process, and identify the structural and dynamics signatures of the toxic oligomers by working closely with experimental groups. The obtained molecular and structural insights to amyloid toxicity will help better understand the disease mechanisms and aid in the design of therapeutic strategies against amyloidosis.
 Our strategy of mitigating amyloid toxicity is to minimize the population of toxic oligomers and/or reduce the direct exposure of cells to these toxic species. The overall population of toxic oligomers can be reduced by stabilizing monomers, non-toxic or “off-pathway” oligomers, or accelerating the formation of non- or less-toxic fibrils. The direct cell exposure to toxic oligomers can also be achieved by caging these culprits. In our lab, we have been exploring the applications of small molecules, endogenous proteins and peptides, and engineered nanomaterials with specific physicochemical properties as amyloid-mitigating agents.
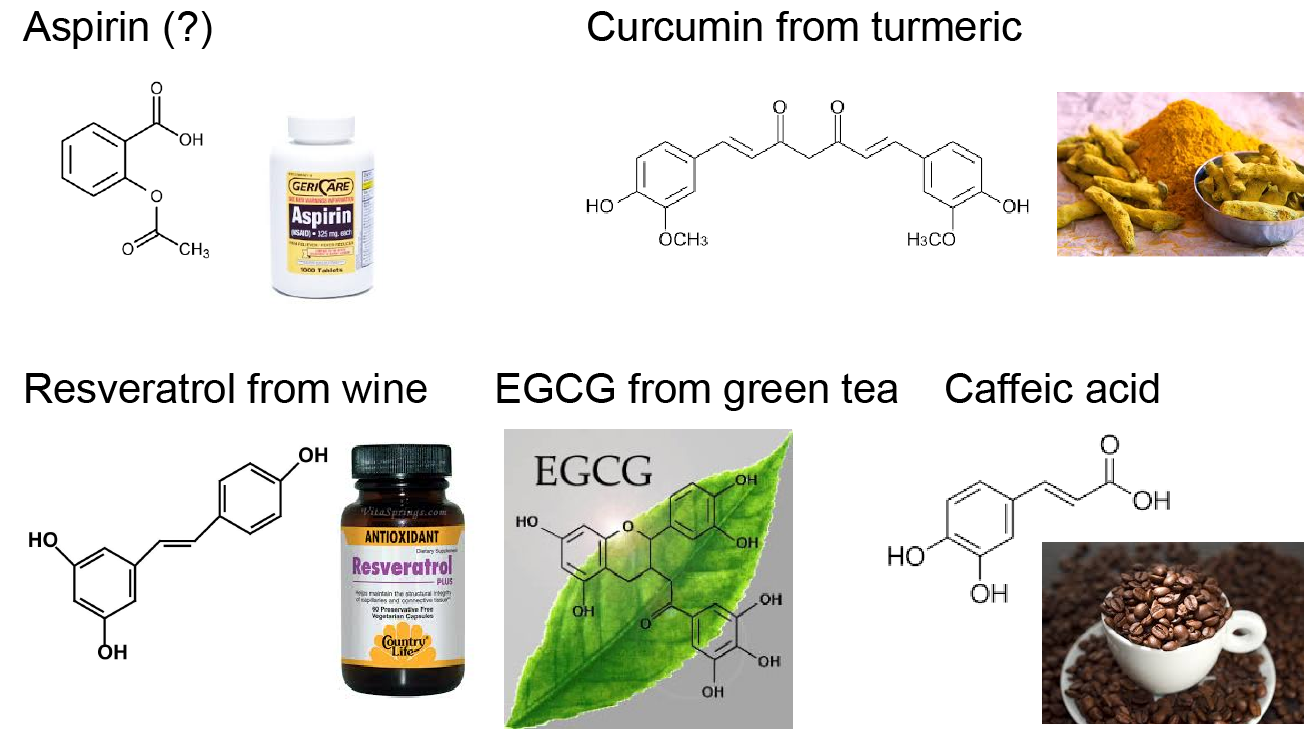
Our strategy of mitigating amyloid toxicity is to minimize the population of toxic oligomers and/or reduce the direct exposure of cells to these toxic species. The overall population of toxic oligomers can be reduced by stabilizing monomers, non-toxic or “off-pathway” oligomers, or accelerating the formation of non- or less-toxic fibrils. The direct cell exposure to toxic oligomers can also be achieved by caging these culprits. In our lab, we have been exploring the applications of small molecules, endogenous proteins and peptides, and engineered nanomaterials with specific physicochemical properties as amyloid-mitigating agents. 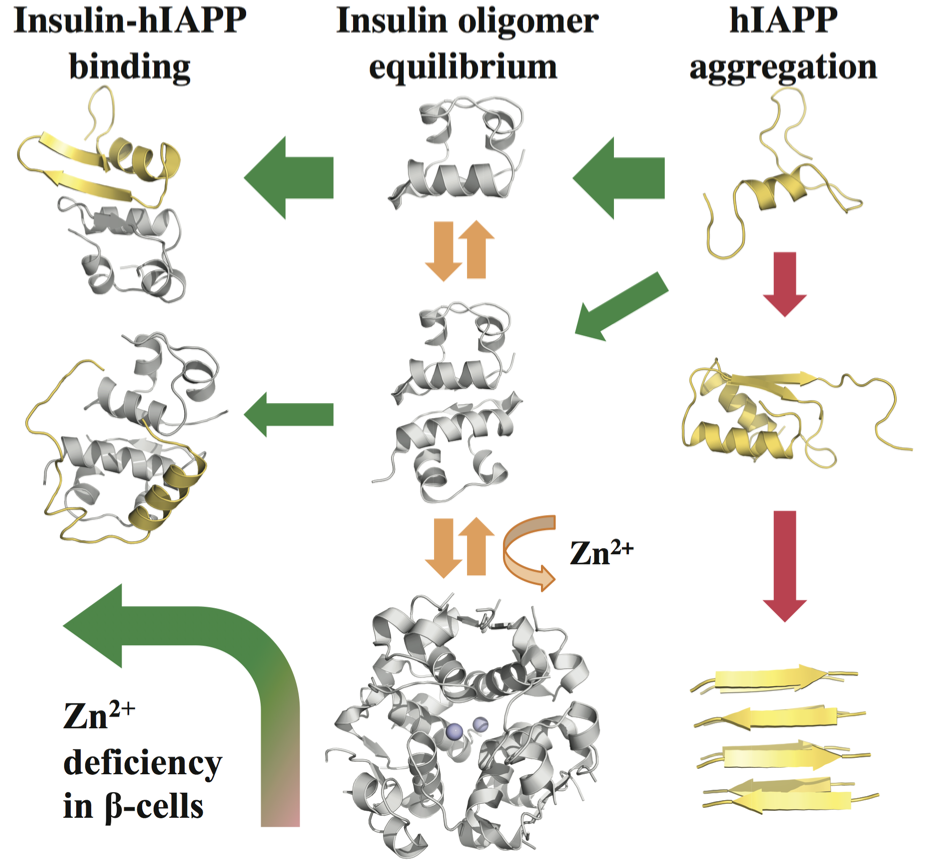 For instance, we have uncovered the amyloid-mitigation mechanisms of several naturally-occurring polyphenols (e.g., resveratrol, curcumin, and EGCG) reported in the literature to be able to inhibit the aggregation of different amyloid peptides. For the aggregation of human islet amyloid polypeptide (hIAPP, aka amylin) associated with beta-cell death in type-2 diabetes, we also studied the effects of various IAPP-colocalizing molecules including insulin, zinc ion, c-peptide and hydrogen ion at low pH in the beta-cell granule, where hIAPP is stored without apparent aggregation for hours between meals at mM concentrations. In the search for anti-amyloid nanomedicine, we have been studying the physicochemical determinants of nanoparticles in mitigating amyloid aggregation and toxicity. All these research projects are highly inter-disciplinary and collaborative, and results from the synergy between simulations and experiments. Molecular modeling and dynamics simulations have been employed not only to help explain experimental characterizations and elucidate molecular mechanisms but also to make predictions and guide experimental validations.
For instance, we have uncovered the amyloid-mitigation mechanisms of several naturally-occurring polyphenols (e.g., resveratrol, curcumin, and EGCG) reported in the literature to be able to inhibit the aggregation of different amyloid peptides. For the aggregation of human islet amyloid polypeptide (hIAPP, aka amylin) associated with beta-cell death in type-2 diabetes, we also studied the effects of various IAPP-colocalizing molecules including insulin, zinc ion, c-peptide and hydrogen ion at low pH in the beta-cell granule, where hIAPP is stored without apparent aggregation for hours between meals at mM concentrations. In the search for anti-amyloid nanomedicine, we have been studying the physicochemical determinants of nanoparticles in mitigating amyloid aggregation and toxicity. All these research projects are highly inter-disciplinary and collaborative, and results from the synergy between simulations and experiments. Molecular modeling and dynamics simulations have been employed not only to help explain experimental characterizations and elucidate molecular mechanisms but also to make predictions and guide experimental validations.
29. Z. Zhang, G. Huang, S. Gupta, E. Sargent, H. Tang, F. Ding, “Determinants for Sub-stoichiometric Inhibition of IAPP and A-Beta Amyloid Aggregations by Bri2 BRICHOS”, ACS Chemical Neuroscience, 16(6): 1150–1160 (2025) doi:10.1021/acschemneuro.4c00839
28. A. Chaari, N. Saikia, P. Paul, M. Yousef, F. Ding, M. Ladjimi, “Experimental and computational investigation of the effect of Hsc70 structural variants on inhibiting amylin aggregation”, Biophysical Chemistry, 309:107235 (2024) doi:10.1016/j.bpc.2024.107235
27. Y. Wang, J. Xu, J. Yan, X. Fan, G. Wei, C. Wang, F. Ding*, Y. Sun*, “SEVI Inhibits Aβ Amyloid Aggregation by Capping the β-Sheet Elongation Edges”, Journal of Chemical Information and Modeling, 63(11):3567-3578 (2023) doi: 10.1021/acs.jcim.3c00414
26. Andrikopoulos N., Li Y., Nandakumar A., Quinn J., Davis T., Ding F.*, Saikia N.*, Ke P.C.*, “Zinc-Epigallocatechin-3-gallate Network-Coated Nanocomposites against the Pathogenesis of Amyloid-Beta”, ACS Applied Materials & Interfaces, 15, 6, 7777–7792 (2023) doi:10.1021/acsami.2c20334
25. N. Benhamou Goldfajn, H. Tang, F. Ding, “Sub-Stoichiometric Inhibition of Insulin against IAPP Aggregation is Attenuated by the Incompletely Processed N-Terminus of proIAPP”, ACS Chemical Neuroscience, 13(13): 2006–2016 (2022) doi:10.1021/acschemneuro.2c00231
24. H. Tang, Y. Li, A. Kakinen, N. Andrikopoulos, Y. Sun, E. Kwak, T. P. Davis, F. Ding* and P. C. Ke*, “Graphene Quantum Dots Obstruct the Membrane Axis of Alzheimer’s Amyloid Beta”, Physical Chemistry Chemical Physics, 24, 86-97 (2022) doi: 10.1039/D1CP04246G
23. N. Andrikopoulos, Z. Song, X. Wan, A. Douek, I. Javed, C. Fu, X. Changkui, Y. Xing, F. Xin, Y. Li, A. Kakinen, K. Koppel, R. Qiao, A. Whittaker, J. Kaslin, T. Davis*, Y. Song*, F. Ding*, P.C. Ke*, “Inhibition of Amyloid Aggregation and Toxicity with Janus Iron Oxide Nanoparticles”, Chem. Mater., 33, 16, 6484–6500 (2021) doi: 10.1021/acs.chemmater.1c01947
22. Y. Li, H. Tang, H. Zhu, A. Kakinen, D. Wang, N. Andrikopoulos, Y. Sun, A. Nandakumar, E. Kwak, T. Davis, D. Leong, F. Ding, P. C. Ke, “Ultrasmall Molybdenum Disulfide Quantum Dots Cage Alzheimer’s Amyloid Beta to Restore Membrane Fluidity”, ACS Appl. Mater. Interfaces, 13(25): 29936–29948 (2021) doi: 10.1021/acsami.1c06478
21. Chen P, Ding F, Cai R, Javed I, Yang W, Zhang Z, Li Y, Davis TP, Ke PC, Chen C., “Amyloidosis Inhibition, a New Frontier of the Protein Corona”, Nano Today, 35:100937 (2020) doi: 10.1016/j.nantod.2020.100937
20. Y. Sun, F. Ding, “αB-Crystallin Chaperone Inhibits Aβ Aggregation by Capping the β-Sheet-Rich Oligomers and Fibrils”, J. Phys. Chem. B, 124:10138-10146 (2020) doi: 10.1021/acs.jpcb.0c07256.
19. Z Huma,I Javed, Z Zhang, H Bilal, Y Sun, SZ Hussain, TP Davis, DE Otzen, CB Landersdorfer, F Ding, I Hussain and PC Ke, “Nano Silver Mitigates Biofilm Formation via FapC Amyloidosis Inhibition”, Small, 1906674-1906683 (2019) DOI: 10.1002/smll.201906674
18. A Faridi, Y Sun, M Mortimer, RR Aranha, A Nandakumar, Y Li, I Javed, A Kakinen, Q Fan, AW Purcell, TP Davis,* F Ding,* P Faridi,* and P Ke*, “Graphene quantum dots rescue protein dysregulation of pancreatic β-cells exposed to human islet amyloid polypeptide”, Nano Research, 12(11), 2827–2834 (2019) DOI: 10.1007/s12274-019-2520-7
17. P.C. Ke, E.H. Pilkington, Y. Sun, I. Javed, A. Kakinen, G. Peng, F. Ding, T.P. Davis, “Mitigation of Amyloidosis with Nanomaterials”, Advanced Materials, 32(18):e1901690, (2020) DOI: 10.1002/adma.201901690
16. M. Wang, Y. Sun, X. Cao, G. Peng, I. Javed, A. Kakinen, T.P. Davis, S. Lin, J. Liu, F. Ding, and P. Ke, “Graphene Quantum Dots against Human IAPP Aggregation and Toxicity in Vivo”, Nanoscale 10, 19995, DOI: 10.1039/C8NR07180B (2018)
15. A. Faridi,Y. Sun, Y. Okazaki, G. Peng, J. Gao, A. Kakinen, P. Faridi, M. Zhao, I. Javed, A.W. Purcell, T.P. Davis, S. Lin, R. Oda, F. Ding, P. Ke, “Mitigating Human IAPP Amyloidogenesis in Vivo with Chiral Silica Nanoribbons”, Small, 14, 1802825, DOI: 10.1002/smll.201802825 (2018)
14. A. Kakinen, J. Adamcik, B. Wang, X. Ge, R. Mezzenga, T.P. Davis, F. Ding, and P. Ke, “Nanoscale inhibition of polymorphic and ambidextrous IAPP amyloid aggregation with small molecules”, Nano Research, 11(7): 3636–3647 (2018)[download] DOI: 10.1007/s12274-017-1930-7
13. Y. Xing, E. H. Pilkington, M. Wang, C. Nowell, A. Kakinen, Y. Sun, B. Wang, T. P. Davis, F. Ding and P. C. Ke, “Lysophosphatidylcholine modulates the aggregation of human islet amyloid polypeptide”, Phys. Chem. Chem. Phys., 19, 30627-30635, 2017, DOI: 10.1039/C7CP06670H
12. E. Pilkington, M. Lai, X. Ge, W. Stanley, B. Wang, M. Wang, A. Kakinen, M. Sani, M. Whittaker, E. Gurzov, F. Ding, J. Quinn, T. Davis, P. Ke, “Star Polymers Reduce IAPP Toxicity via Accelerated Amyloid Aggregation”, Biomacromolecules, 18 4249–4260, (2017) DOI: 10.1021/acs.biomac.7b01301
11. I. Javed, Y. Sun, J. Adamcik, B. Wang, A. Kakinen, E. Pilkington, F. Ding, R. Mezzenga, T. Davis, Thomas; P. Ke, “Co-fibrillization of pathogenic and functional amyloid proteins with gold nanoparticles against amyloidogenesis”, Biomacromolecules, 18, 4316–4322 (2017) DOI: 10.1021/acs.biomac.7b01359
10. B. Wang, E.H. Pilkington, Y. Sun, T.P. Davis, P.C. Ke and F. Ding, “Modulating protein amyloid aggregation with nanomaterials”, Environmental Science Nano, 4, 1772-1783 (2017) DOI: 10.1039/C7EN00436B
9. X. Ge, A. Kakinen, E.N. Gurzov, W. Yang, L. Pang, E.H. Pilkington, P. Govindan-Nedumpully, P. Chen, F. Separovic, T.P. Davis, P. C. Ke, and F. Ding, “Zinc-coordination and C-peptide complexation: a potential mechanism for the endogenous inhibition of IAPP aggregation”, Chem. Comm., 53(68):9394-9397 (2017) DOI: 10.1039/C7CC04291D
8. Pilkington E.H., Xing Y., Wang B., Kakinen A., Wang M., Davis T.P., Ding F., Ke P.C., “Effects of Protein Corona on IAPP Amyloid Aggregation, Fibril Remodelling, and Cytotoxicity”, Scientific Reports, 7(1):2455 (2017)
7. E.N. Gurzov, B. Wang, E.H. Pilkington, P. Chen,A. Kakinen, W.J. Stanley, S.A. Litwak, E.G. Hanssen,T.P. Davis, F. Ding, and P.Chun Ke, “Inhibition of hIAPP Amyloid Aggregation and Pancreatic β-cell Toxicity by OH-terminated PAMAM Dendrimer”, Small, 12(12):1615–1626 (2016)
6. P. Nedumpully-Govindan, E.N. Gurzov, P. Chen, E.H. Pilkington, W.J. Stanley, S.A. Litwak, T.P. Davis, P.C. Ke, and F. Ding, “Graphene Oxide Inhibits hIAPP Amyloid Fibrillation and Toxicity in Insulin-Producing NIT-1 Cells”, Phys. Chem. Chem. Phys., 18:94-100 (2016)
5. P. Nedumpully-Govindan, A. Kakinen, E.H. Pilkington, T.P. Davis, P.C. Ke and F. Ding, “Stabilizing Off-pathway Oligomers by Polyphenol Nanoassemblies for IAPP Aggregation Inhibition”, Scientific Reports 6: 19463 (2016)
4. S. Radic, T.P. Davis, P.C. Ke and F. Ding, “Contrasting effects of nanoparticle-protein attraction on amyloid aggregation”, RSC Advances, 5, 105489-105498 (2015)
3. P. Nedumpully-Govindan, Y. Yang, R. Andorfer, W. Cao, and F. Ding, “Promotion or Inhibition of IAPP Aggregation by Zinc Coordination Depends on Its Relative Concentration”, Biochemistry, 54:7335-44 (2015)
2. Nedumpully-Govindan P. and Ding F., “Inhibition of IAPP aggregation by insulin depends on the insulin oligomeric state regulated by zinc ion concentration”, Scientific Reports 5, (2015)
1. S. Radic, P. Nedumpully-Govindan,R. Chen, E. Salonen, J.M. Brown, P.C. Ke, and F. Ding, “Effect of Fullerenol Surface Chemistry on Nanoparticle Binding-induced Protein Misfolding”, Nanoscale, 6 (14), 8340 – 8349 (2014)
