A major challenge in modeling molecular recognition is the conformational flexibility. The structures of the receptors in the bound and un-bound states are often different, known as the induced-fit problem. In these cases, a rigid docking approach will fail to achieve accurate predictions. Existing flexible methods for modeling molecular recognition or docking often adopt the ensemble docking approach, where an ensemble of receptor and/or ligand conformations are pre-constructed. Rigid or semi-rigid docking is performed onto these structures in the ensemble. The predicted binding poses are then selected from the grand ensemble of poses. In another words, the conformational flexibility of ligands and receptors are modeled in a decoupled or loose-coupled manner. Once the bound-conformation is not included in the pre-constructed ensemble, accurate prediction will be difficult. Alternatively, we fully integrate the conformational flexibility into the modeling of molecular recognition.
A. MedusaDock–a flexible docking approach. MedusaDock models both ligand and receptor flexibility simultaneously with sets of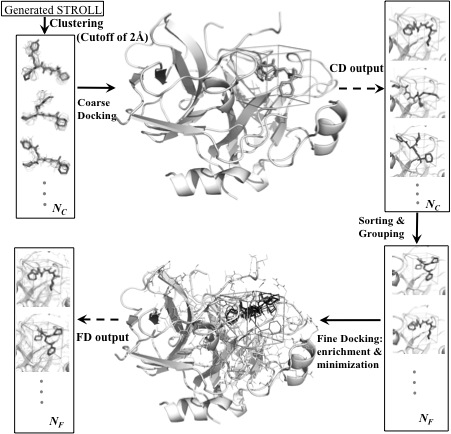 discrete rotamers. We developed an algorithm to build the ligand rotamer library “on-the-fly” during docking simulations. MedusaDock benchmarks demonstrate a rapid sampling efficiency and high prediction accuracy in both self- (to the co-crystallized state) and cross-docking (to a state co-crystallized with a different ligand), the latter of which mimics the virtual screening procedure in computational drug discovery. We also perform a virtual screening test of four flexible kinase targets, including cyclin-dependent kinase 2, vascular endothelial growth factor receptor 2, HIV reverse transcriptase, and HIV protease. We find significant improvements of virtual screening enrichments when compared to rigid-receptor methods. The predictive power of MedusaDock in cross-docking and preliminary virtual-screening benchmarks highlights the importance to model both ligand and receptor flexibility simultaneously in computational docking.
discrete rotamers. We developed an algorithm to build the ligand rotamer library “on-the-fly” during docking simulations. MedusaDock benchmarks demonstrate a rapid sampling efficiency and high prediction accuracy in both self- (to the co-crystallized state) and cross-docking (to a state co-crystallized with a different ligand), the latter of which mimics the virtual screening procedure in computational drug discovery. We also perform a virtual screening test of four flexible kinase targets, including cyclin-dependent kinase 2, vascular endothelial growth factor receptor 2, HIV reverse transcriptase, and HIV protease. We find significant improvements of virtual screening enrichments when compared to rigid-receptor methods. The predictive power of MedusaDock in cross-docking and preliminary virtual-screening benchmarks highlights the importance to model both ligand and receptor flexibility simultaneously in computational docking.
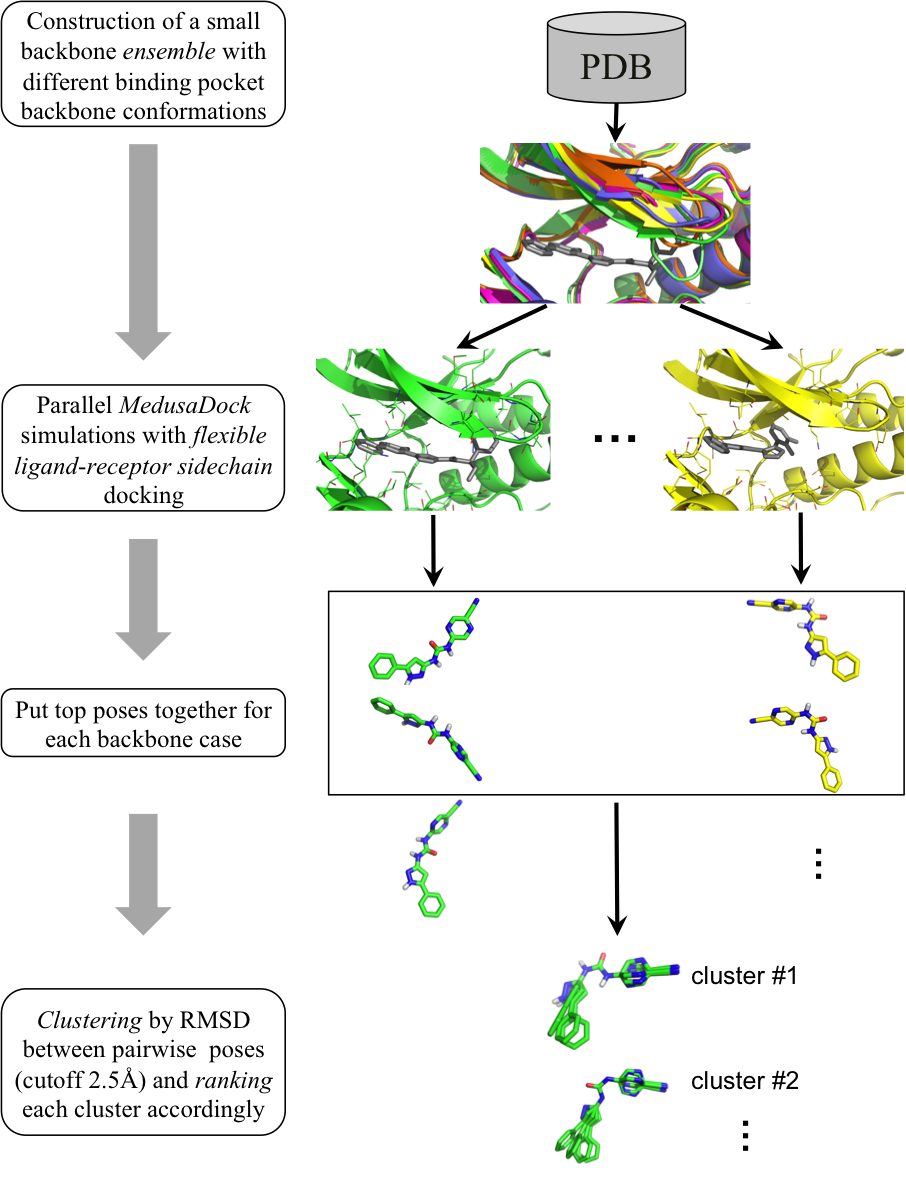 B. Incorporating backbone flexibility in MedusaDock. We introduce backbone flexibility into MedusaDock by implementing ensemble docking in a sequential manner for a set of distinct receptor backbone conformations. We generate corresponding backbone ensembles to capture backbone changes upon binding to different ligands, as observed experimentally. We develop a simple clustering and ranking approach to select the top poses as blind predictions. We applied our method in the CSAR2011 benchmark exercise. In 28 out of 35 cases (80%) where the ligand-receptor complex structures were released, we were able to predict near-native poses (< 2.5 Å RMSD), the highest success rate reported for CSAR2011. Thisresult highlights the importance of modeling receptor backbone flexibility to the accurate docking of ligands to flexible targets. We expect a broach application of our fully flexible docking approach in biological studies as well as in rational drug design.
B. Incorporating backbone flexibility in MedusaDock. We introduce backbone flexibility into MedusaDock by implementing ensemble docking in a sequential manner for a set of distinct receptor backbone conformations. We generate corresponding backbone ensembles to capture backbone changes upon binding to different ligands, as observed experimentally. We develop a simple clustering and ranking approach to select the top poses as blind predictions. We applied our method in the CSAR2011 benchmark exercise. In 28 out of 35 cases (80%) where the ligand-receptor complex structures were released, we were able to predict near-native poses (< 2.5 Å RMSD), the highest success rate reported for CSAR2011. Thisresult highlights the importance of modeling receptor backbone flexibility to the accurate docking of ligands to flexible targets. We expect a broach application of our fully flexible docking approach in biological studies as well as in rational drug design.
C. Protein-peptide binding using DMD simulations. Utilizing 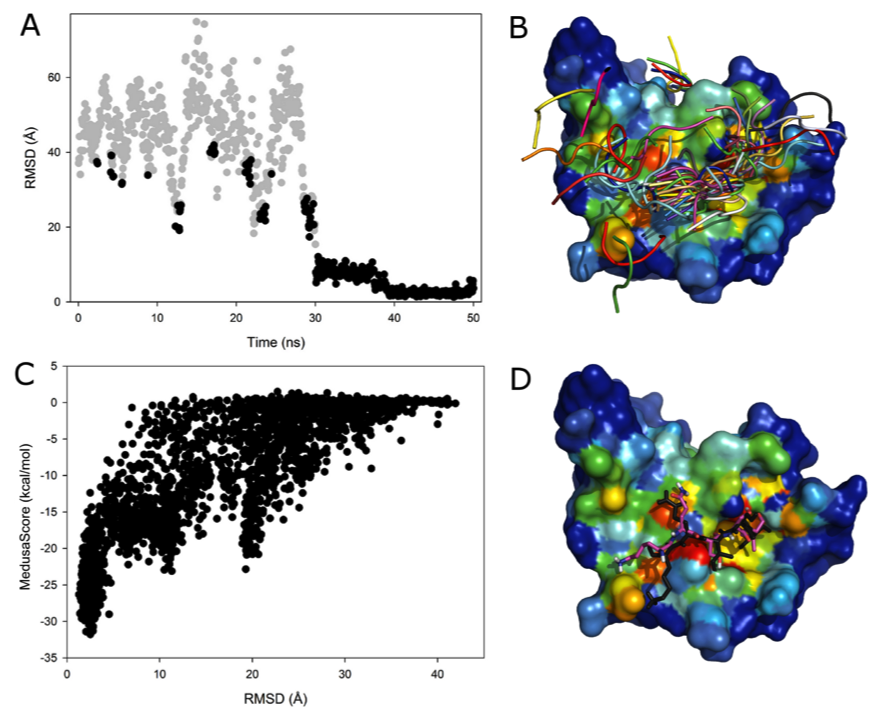 sampling efficiency of DMD and Medusa force field in describing interaction between amino acids, we apply DMD simulations to modeling protein-peptide recognition. We find that, in most cases, we recapitulate the native binding sites and native-likeposes of protein-peptide complexes. Inclusion ofelectrostatic interactions in simulations significantly improves the prediction accuracy. Our results alsohighlight the importance of protein conformationalflexibility, especially side-chain movement, which allows the peptide to optimize its conformation. Ourfindings not only demonstrate the importance of sufficient sampling of the protein and peptide conformations, but also reveal the possible effects ofelectrostatics and conformational flexibility on peptide recognition.
sampling efficiency of DMD and Medusa force field in describing interaction between amino acids, we apply DMD simulations to modeling protein-peptide recognition. We find that, in most cases, we recapitulate the native binding sites and native-likeposes of protein-peptide complexes. Inclusion ofelectrostatic interactions in simulations significantly improves the prediction accuracy. Our results alsohighlight the importance of protein conformationalflexibility, especially side-chain movement, which allows the peptide to optimize its conformation. Ourfindings not only demonstrate the importance of sufficient sampling of the protein and peptide conformations, but also reveal the possible effects ofelectrostatics and conformational flexibility on peptide recognition.
5. Nedumpully-Govindan P., Domen J., and Ding F., “CSAR Benchmark of flexible MedusaDock in affinity prediction and native-like binding pose selection”, Journal of Chemical Information and Modeling, in press (2015)
4. Fourche, D., Muratov, E., Ding, F., Dokholyan, N. V., Tropsha, A. “Predicting binding affinity of CSAR ligands using both structure-based and ligand-based approaches”, Journal of Chemical Information and Modeling, in press (2013)
3. F. Ding. and Dokholyan, N. V., “Incorporating backbone flexibility in MedusaDock improves ligand binding pose prediction in the CSAR2011 docking benchmark”, Journal of Chemical Information and Modeling, in press, (2012)[download]
2. Dagliyan, O., Proctor, E. A., D’Auria, K., F. Ding* and Dokholyan N. V.* “Structural and Dynamic Determinants of Protein-peptide Recognition”, Structure, 19:1837 (2011) [download]
1. F. Ding, Yin, S., and Dokholyan, N. V. “Rapid flexible docking using a stochastic rotamer library of ligands”, Journal of Chemical Information and Modeling, 50:1623-32 (2010)[download]
